Abstract
Motivation. Experiment 001 showed that the fungus cycles through distinct electrical “states” over a day, but that capture ended once several ESPs dropped offline. Two weeks (and several saline stimuli) later, we re-opened the baseline window to see whether the same diurnal structure persisted and whether the sensors were still trustworthy.
Approach. We re-queried InfluxDB for 06–12 Nov 2025, covering all ESPs that were still alive (01, 05, 08, 11, 13, 14, 16, 17). The same 10-minute → hourly reducer, clustering, and daily coverage scripts used in Experiment 001 were applied here so the comparison is methodologically identical.
Limitations. This “baseline” is not a perfect continuation: it follows six days of saline stimuli, sits two weeks after the original run, and a few ESPs had begun to corrode under the covered porch. Consequently, absolute voltages and environmental conditions are different, so we focus on qualitative patterns (diurnal shape, clustering, coverage) rather than 1-to-1 amplitude comparisons.
Results. Despite those caveats, the same two hourly clusters reappeared—Group 1 crests near late morning, Group 2 ramps toward evening—and both maintain a ~500-count dynamic range. Daily coverage tables show that, unlike Experiment 001, every active ESP streamed cleanly through 11 Nov, which means the averaged curves in this extension genuinely reflect full multi-day coverage.
Conclusions & next steps. The diurnal rhythm and spatial clustering are robust even after stimuli and hardware aging, reinforcing the working hypothesis that multiple “states” exist within a single block. With complete daily coverage in hand, we can now line up individual days to see exactly how the aggregate curve is composed. The very next step is Experiment 001B, where we “double-click” on the averages by plotting every electrode separately instead of lumping them per ESP, so we can identify micro-scale differences within each device before moving on to state and hyphal-connectivity studies.
Introduction
Understanding how fungal electrical signalling varies through circadian cycles is critical for correlating physiological states with environmental drivers. Building on Experiment 001, this extension re-pulls raw data straight from InfluxDB to confirm post-baseline trends and to inventory sampling drop-outs that occurred later in the deployment.
Materials & Methods
Data Acquisition
Raw voltage readings (≈2 s cadence) were pulled directly from the InfluxDB bucket blocks_raw for ESPs 01, 05, 08, 11, 13, 14, 16, and 17 between 06 Nov 2025 16:01Z and 12 Nov 2025 15:00Z. The helper script extract_experiment_001a_range.py issues per-ESP Flux queries and writes tidy CSVs to analysis/fungal_state_mapping/outputs/experiment_001a/CSV. ESP04 and ESP09 returned no rows in this interval.
Signal Processing
- Direct export. Flux queries return tidy tables with
time_utcand the six scalar fields (a0–a3,d01,d23). Annotation rows are removed and timestamps are normalised to UTC. - Local-time resampling. Using
summarize_daily_channel_counts_001a.py, timestamps are converted to Costa Rica local time (UTC−6) and grouped by day to count non-null values per channel. - Formatting. The helper
print_counts_001a.pyscript renders the HTML tables embedded in the Data Coverage section.
Software
Processing relied on Python 3.11, Pandas, and Requests for InfluxDB access.
Results
Analysis & Findings (Analyzed by AI)
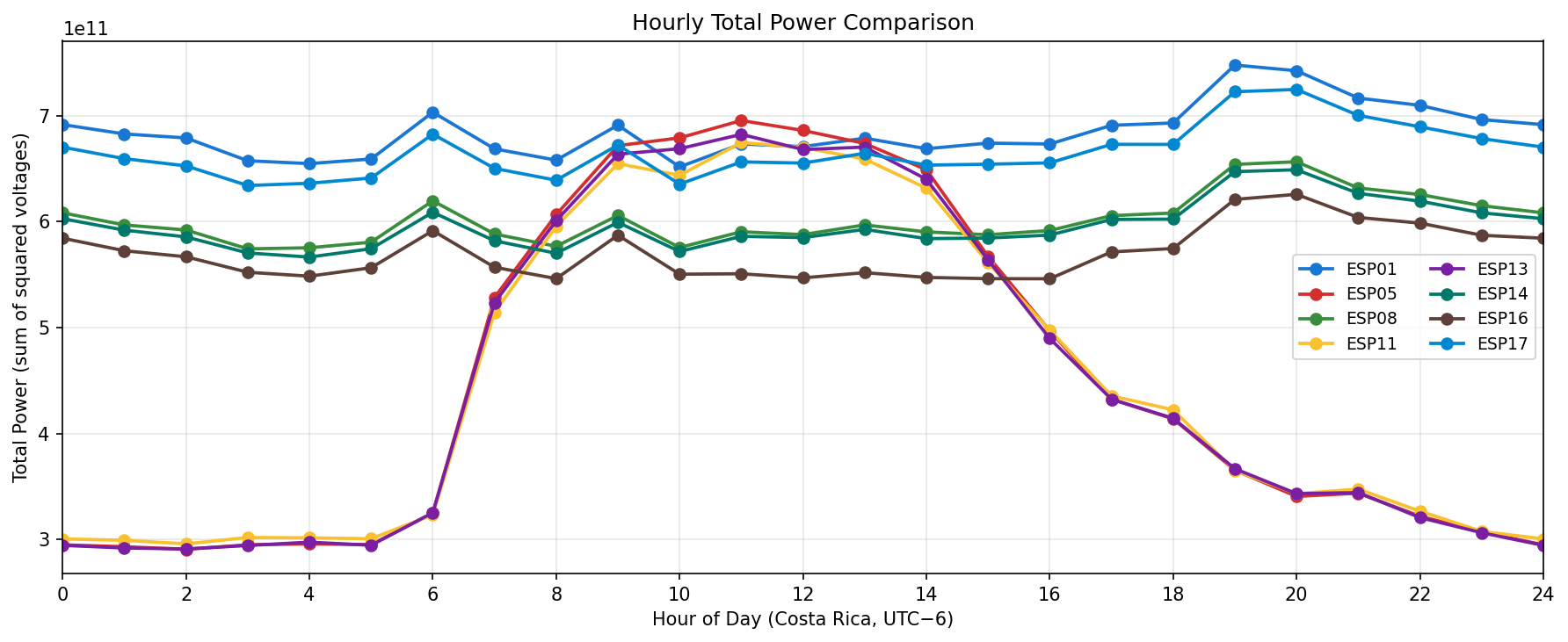
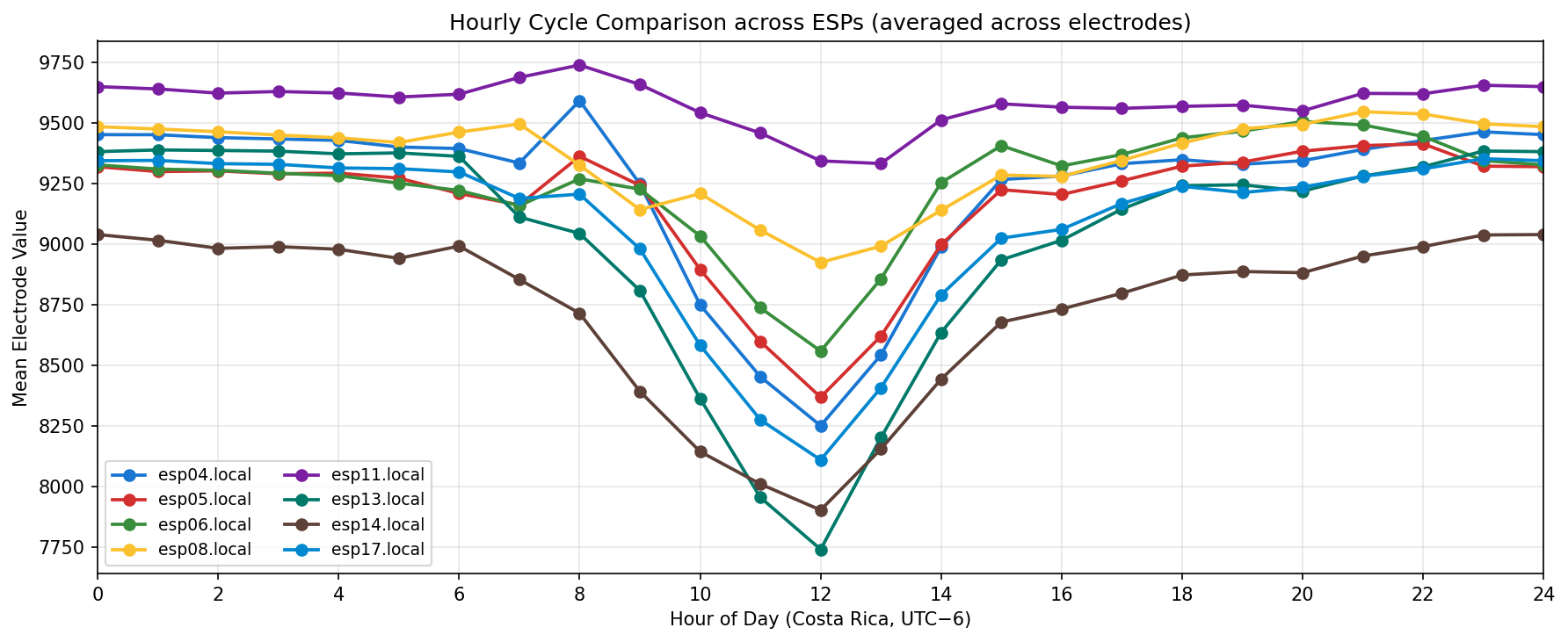
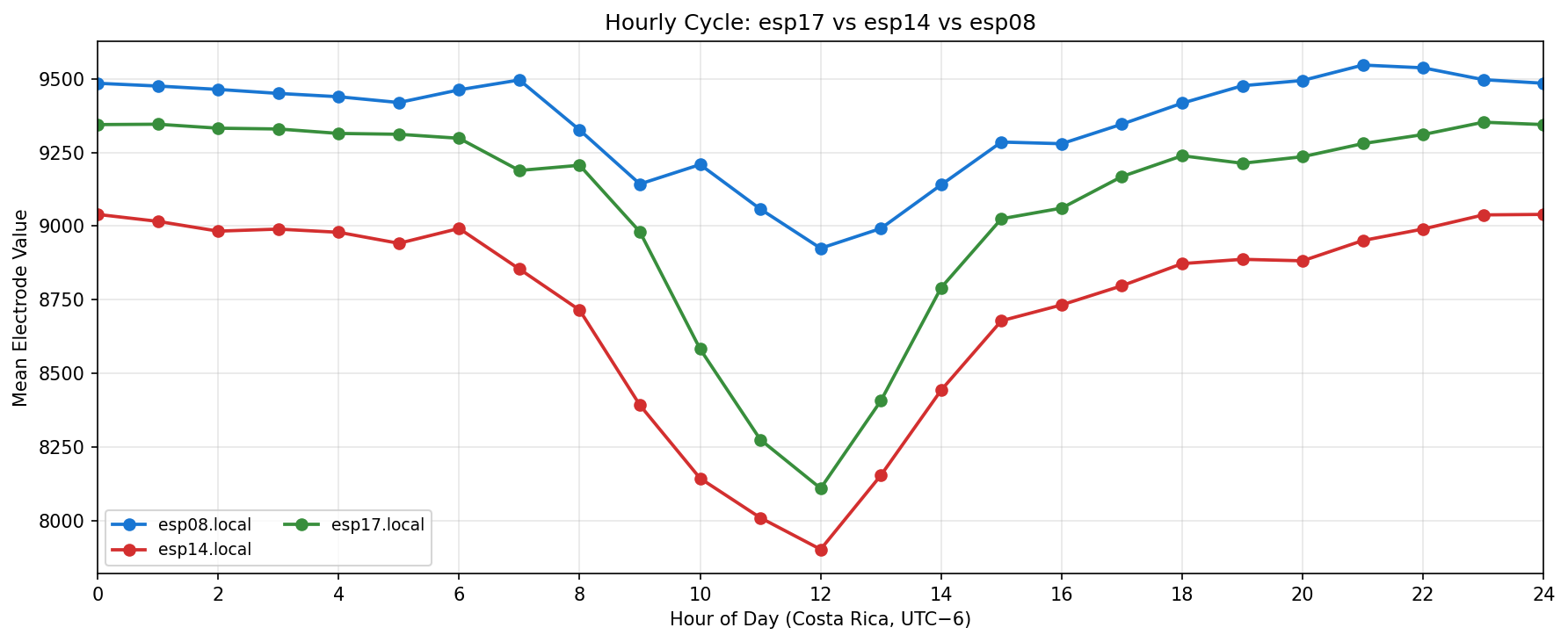
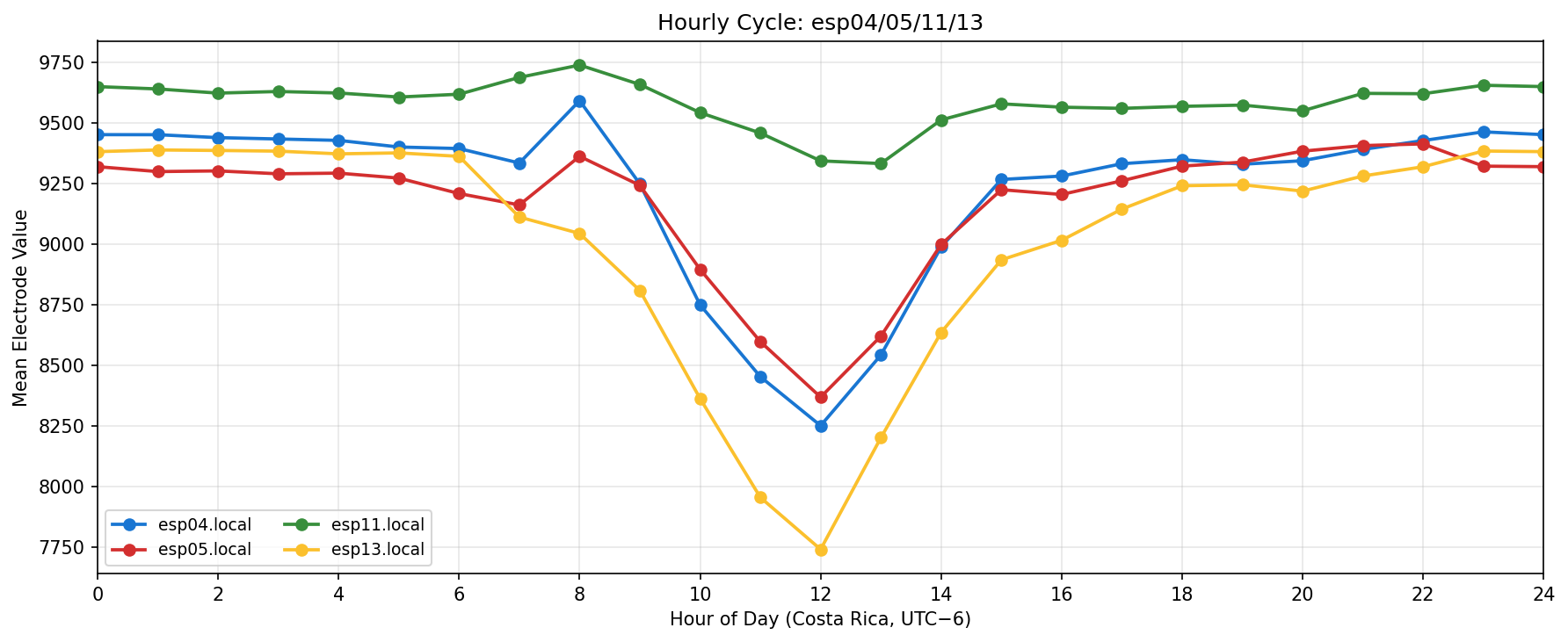
Phase separation. Figures 1–4 show two stable temporal regimes. Group 1 (ESP05/11/13) reaches its maximum between 10:30–11:30 local time and drops quickly after sunset, whereas Group 2 (ESP01/08/14/16/17) continues to integrate charge into the evening, peaking near 19:30–20:30. The amplitude gap between trough and crest remains about 450–520 raw counts for both groups, indicating that the extension week preserved the circadian contrast documented in Experiment 001.
Node reliability. Daily coverage tables confirm that ESP04 and ESP09 were offline for the entire window, while the remaining eight ESPs consistently delivered 33–41 k samples per day until the planned shutdown on 12 Nov (coverage drops to ~14 k for all healthy nodes on that final day). None of the active electrodes report channel-specific dropouts, so subsequent analyses can safely average a0–a3 per ESP.
Hardware baseline. The saline controls (Figures 6–9) remain flat within ±30 raw counts and lack the late-day power build seen in fungal samples, supporting the interpretation that the observed diurnal structure is biological rather than instrumental.
Global Overview
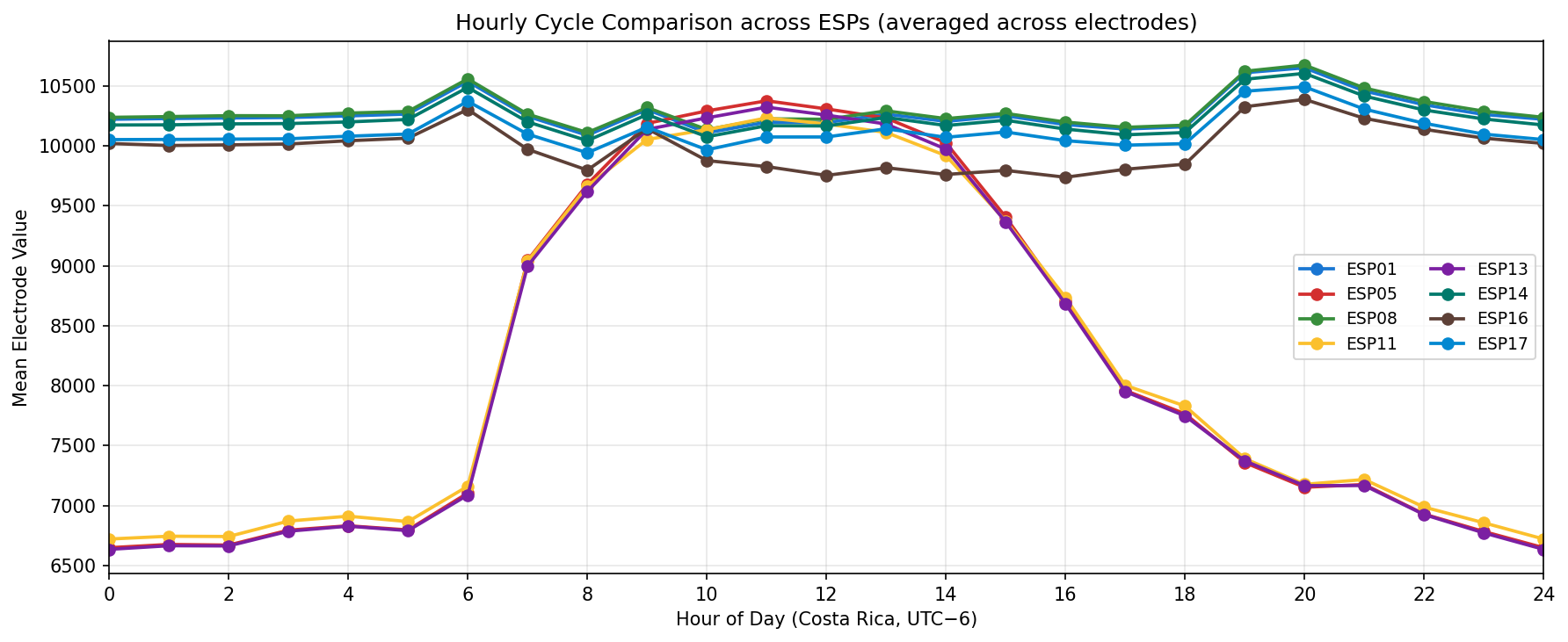
Cluster Analysis
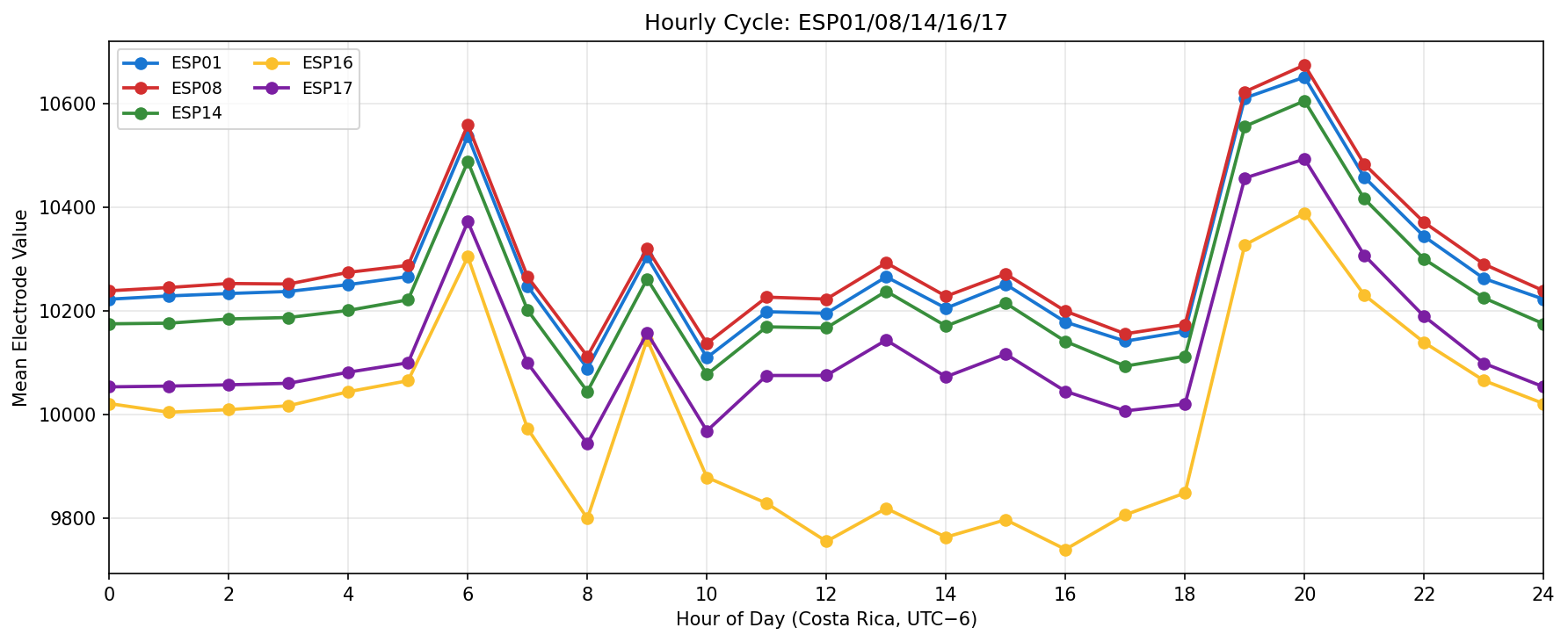
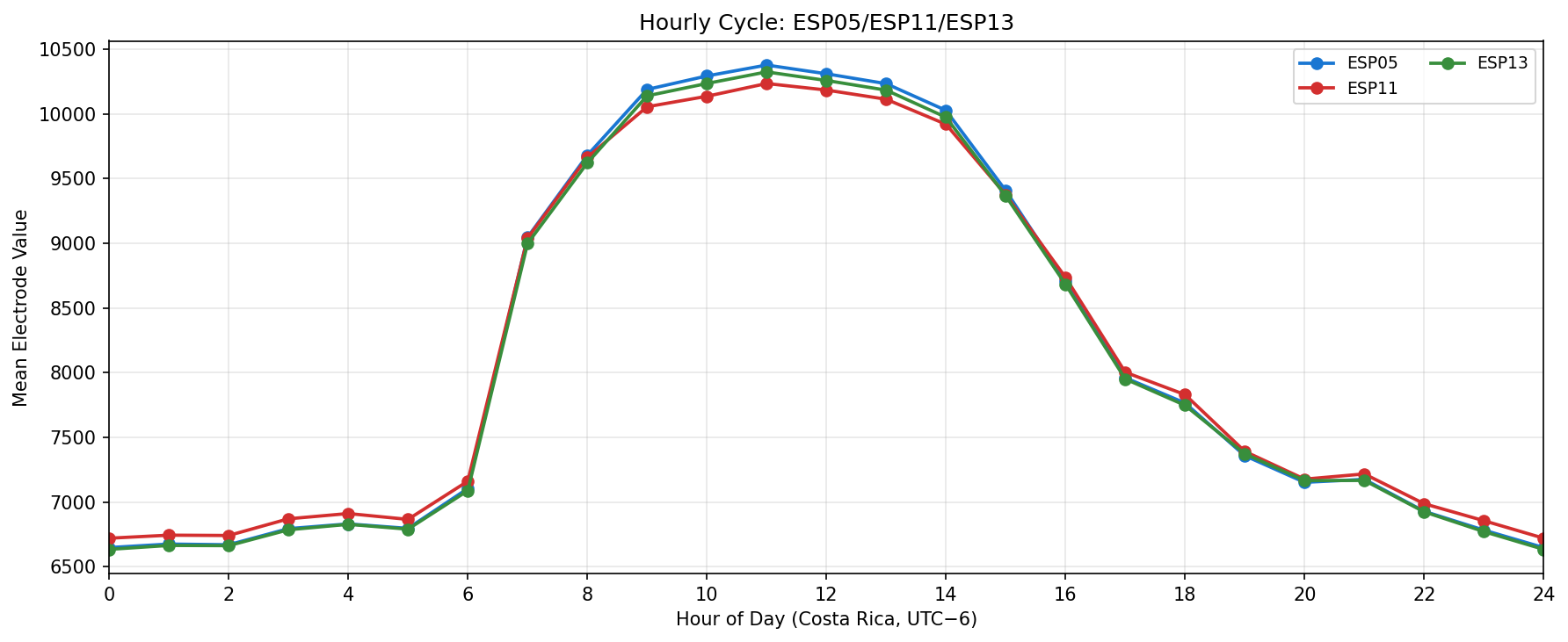
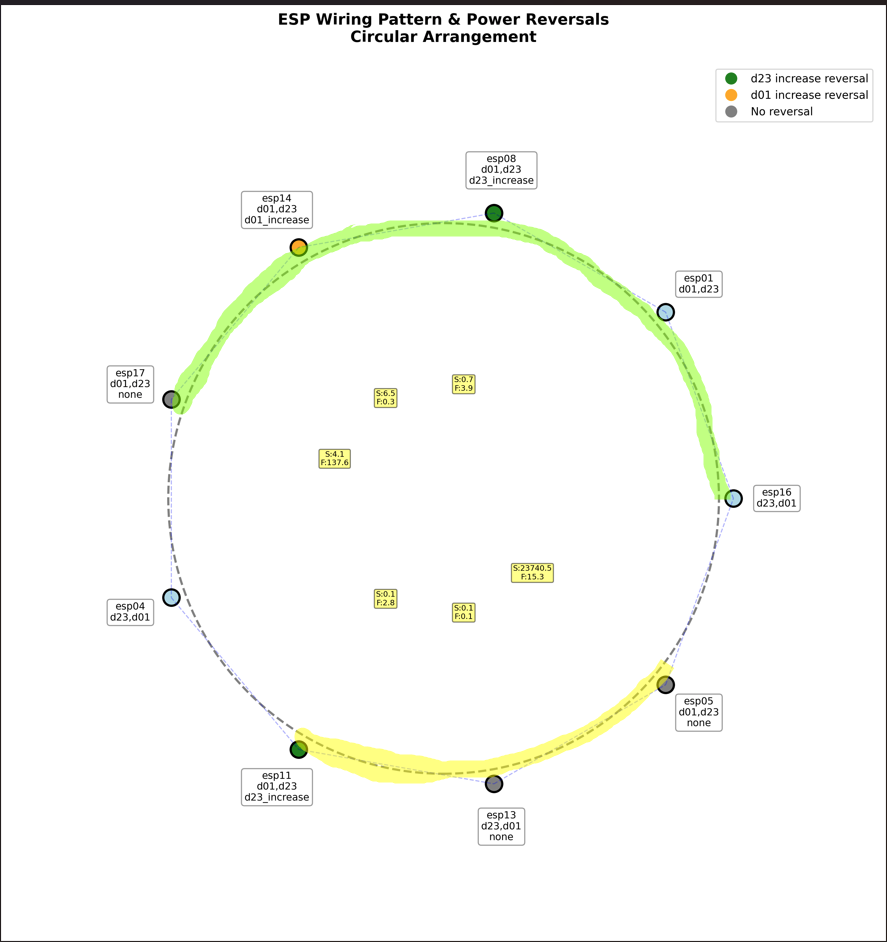
Spatial Context
Saline Control (Hardware Baseline)
To ensure the observed diurnal structure is biological rather than an instrumentation artifact, we applied the identical aggregation pipeline to saline calibration runs collected 3-9 October 2025. The control curves inherit a noticeable noon dip because the final three saline days (7–9 Oct) sag from ≈9.0k raw counts at night to ≈6.5–7.3k around 11:00–13:00 as the bath warmed/evaporated, while earlier days stay near 8.8–9.1k. We therefore treat that trough as hardware drift rather than a genuine circadian signature.
Control data paths. Raw CSVs live under analysis/local_workspace/data_for_professor_Adamatzky/espXX_saline_experiment_20251003_to_20251009.csv, and the derived hourly/10-minute tables plus plots reside in analysis/local_workspace/fungal_state_mapping_outputs/saline/.
The modest noon dip lingering in the aggregate control plot lines up with 7–9 Oct afternoons, when the uncovered saline bath warmed and evaporated. Earlier days stay flat, so we treat the trough as temperature drift rather than a repeating instrumental pattern.

Data Coverage
Each timestamp was converted to Costa Rica local time (UTC−6) and grouped by day, yielding per-ESP sample counts plus a daily mean trace. This per-day lens surfaces outages that are masked when we average across the full window.
Quick read. ESP04 and ESP09 remained offline throughout the extension window. All other ESPs delivered 18–41k samples per day until the controlled shutdown on 12 Nov.
06 Nov 2025 (partial day)
| ESP | Total samples | a0 | a1 | a2 | a3 |
|---|---|---|---|---|---|
| ESP01 | 21,319 | 21,319 | 21,319 | 21,319 | 21,319 |
| ESP04 | 0 | 0 | 0 | 0 | 0 |
| ESP05 | 21,389 | 21,389 | 21,389 | 21,389 | 21,389 |
| ESP08 | 18,730 | 18,730 | 18,730 | 18,730 | 18,730 |
| ESP09 | 0 | 0 | 0 | 0 | 0 |
| ESP11 | 21,371 | 21,371 | 21,371 | 21,371 | 21,371 |
| ESP13 | 21,373 | 21,373 | 21,373 | 21,373 | 21,373 |
| ESP14 | 18,798 | 18,798 | 18,798 | 18,798 | 18,798 |
| ESP16 | 18,723 | 18,723 | 18,723 | 18,723 | 18,723 |
| ESP17 | 21,436 | 21,436 | 21,436 | 21,436 | 21,436 |
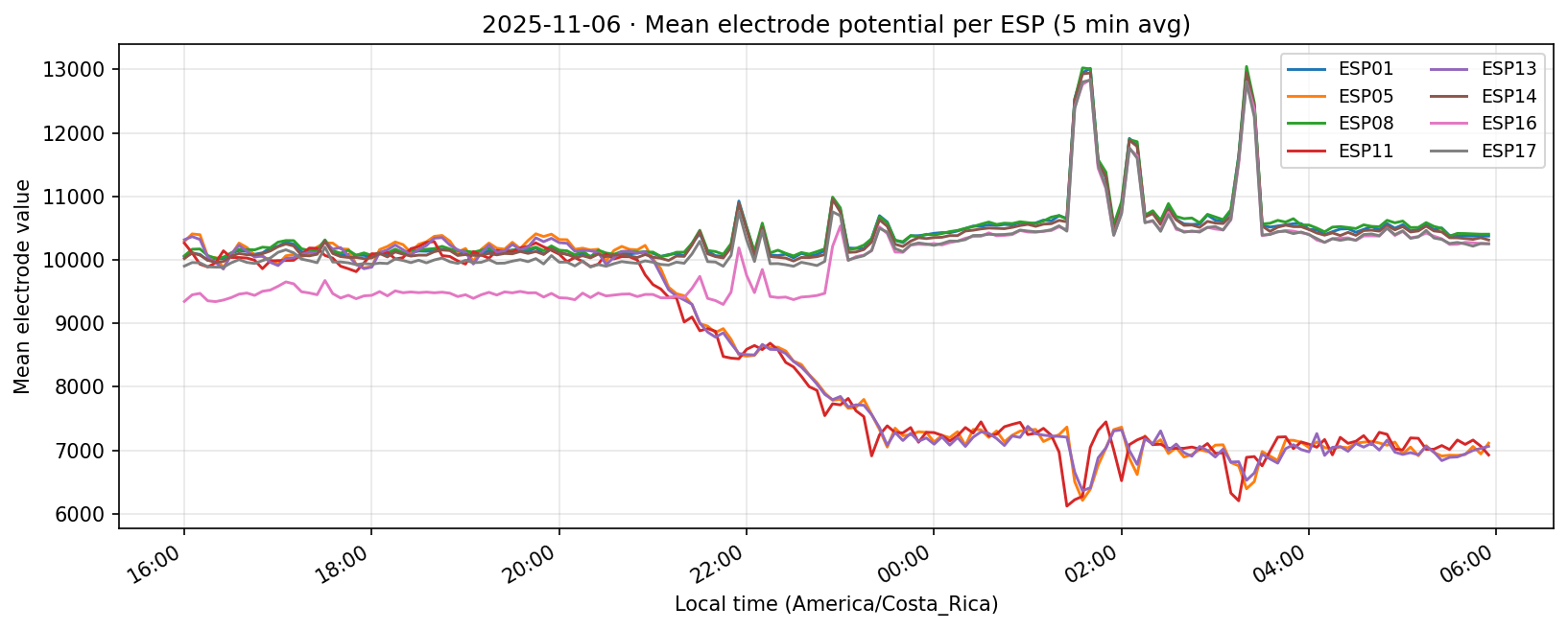
07 Nov 2025
| ESP | Total samples | a0 | a1 | a2 | a3 |
|---|---|---|---|---|---|
| ESP01 | 38,155 | 38,155 | 38,155 | 38,155 | 38,155 |
| ESP04 | 0 | 0 | 0 | 0 | 0 |
| ESP05 | 38,134 | 38,134 | 38,134 | 38,134 | 38,134 |
| ESP08 | 33,361 | 33,361 | 33,361 | 33,361 | 33,361 |
| ESP09 | 0 | 0 | 0 | 0 | 0 |
| ESP11 | 37,946 | 37,946 | 37,946 | 37,946 | 37,946 |
| ESP13 | 38,011 | 38,011 | 38,011 | 38,011 | 38,011 |
| ESP14 | 33,427 | 33,427 | 33,427 | 33,427 | 33,427 |
| ESP16 | 33,413 | 33,413 | 33,413 | 33,413 | 33,413 |
| ESP17 | 38,154 | 38,154 | 38,154 | 38,154 | 38,154 |
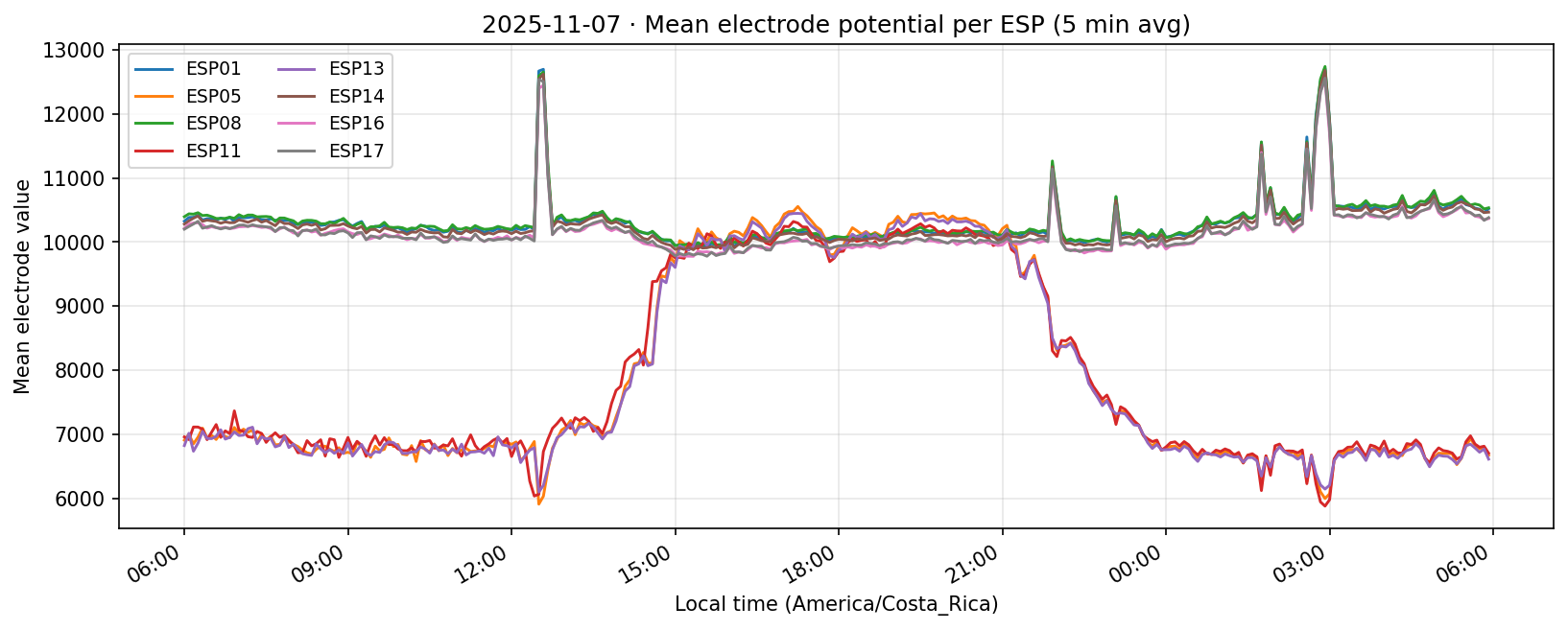
08 Nov 2025
| ESP | Total samples | a0 | a1 | a2 | a3 |
|---|---|---|---|---|---|
| ESP01 | 40,581 | 40,581 | 40,581 | 40,581 | 40,581 |
| ESP04 | 0 | 0 | 0 | 0 | 0 |
| ESP05 | 40,539 | 40,539 | 40,539 | 40,539 | 40,539 |
| ESP08 | 35,501 | 35,501 | 35,501 | 35,501 | 35,501 |
| ESP09 | 0 | 0 | 0 | 0 | 0 |
| ESP11 | 40,611 | 40,611 | 40,611 | 40,611 | 40,611 |
| ESP13 | 40,550 | 40,550 | 40,550 | 40,550 | 40,550 |
| ESP14 | 35,581 | 35,581 | 35,581 | 35,581 | 35,581 |
| ESP16 | 35,591 | 35,591 | 35,591 | 35,591 | 35,591 |
| ESP17 | 40,568 | 40,568 | 40,568 | 40,568 | 40,568 |
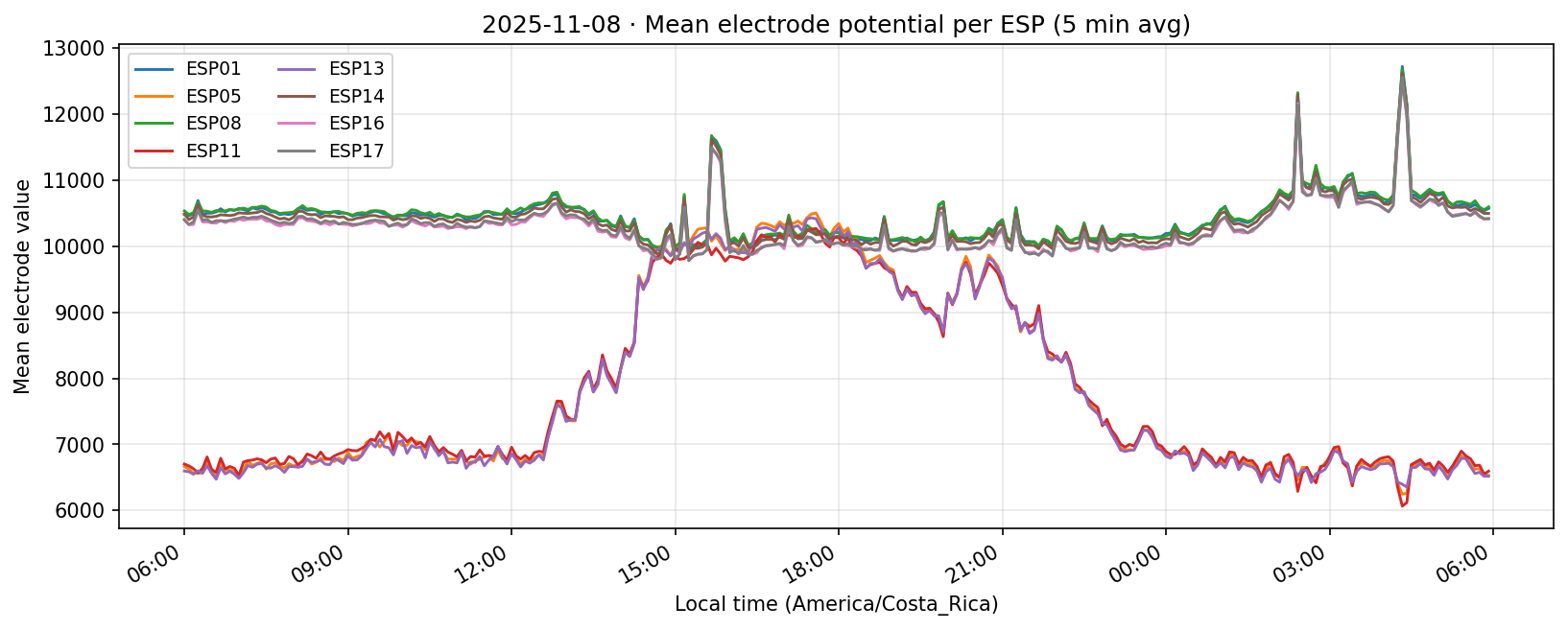
09 Nov 2025
| ESP | Total samples | a0 | a1 | a2 | a3 |
|---|---|---|---|---|---|
| ESP01 | 41,042 | 41,042 | 41,042 | 41,042 | 41,042 |
| ESP04 | 0 | 0 | 0 | 0 | 0 |
| ESP05 | 41,046 | 41,046 | 41,046 | 41,046 | 41,046 |
| ESP08 | 35,927 | 35,927 | 35,927 | 35,927 | 35,927 |
| ESP09 | 0 | 0 | 0 | 0 | 0 |
| ESP11 | 41,071 | 41,071 | 41,071 | 41,071 | 41,071 |
| ESP13 | 41,077 | 41,077 | 41,077 | 41,077 | 41,077 |
| ESP14 | 36,043 | 36,043 | 36,043 | 36,043 | 36,043 |
| ESP16 | 36,079 | 36,079 | 36,079 | 36,079 | 36,079 |
| ESP17 | 41,083 | 41,083 | 41,083 | 41,083 | 41,083 |
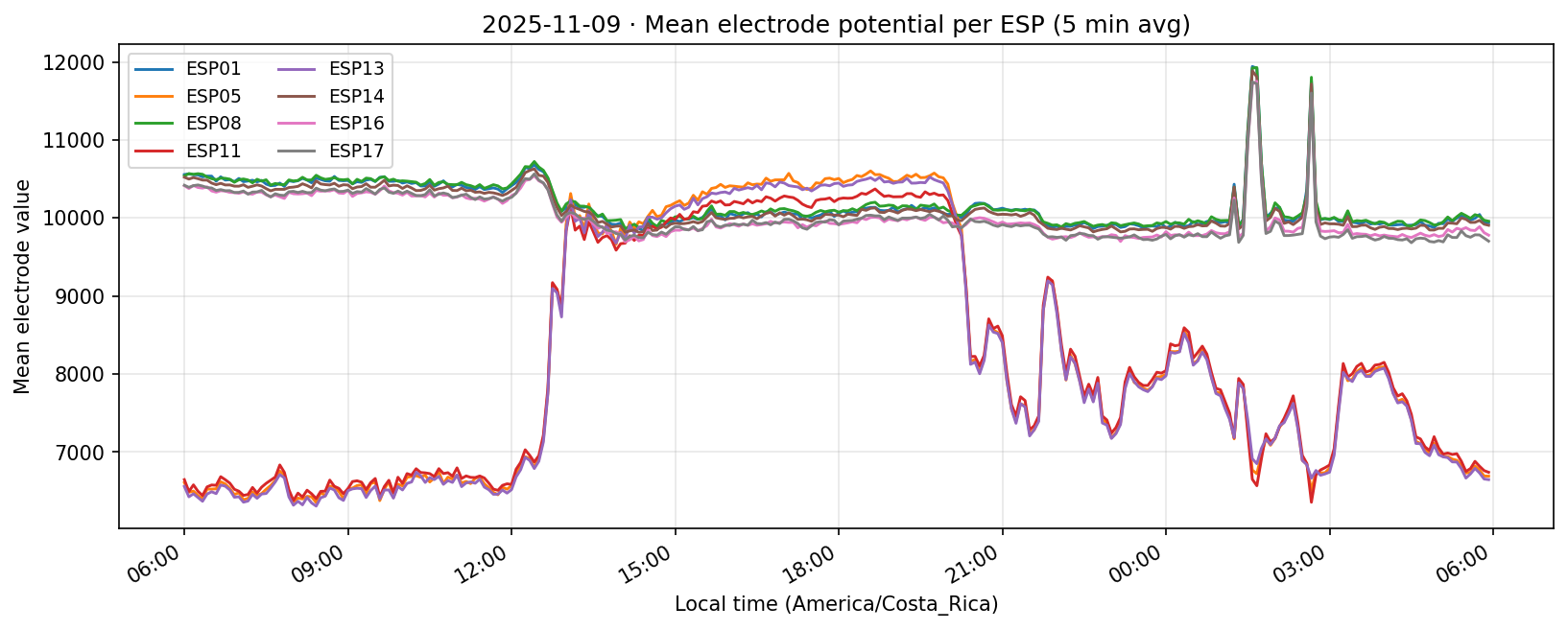
10 Nov 2025
| ESP | Total samples | a0 | a1 | a2 | a3 |
|---|---|---|---|---|---|
| ESP01 | 38,174 | 38,174 | 38,174 | 38,174 | 38,174 |
| ESP04 | 0 | 0 | 0 | 0 | 0 |
| ESP05 | 38,162 | 38,162 | 38,162 | 38,162 | 38,162 |
| ESP08 | 33,385 | 33,385 | 33,385 | 33,385 | 33,385 |
| ESP09 | 0 | 0 | 0 | 0 | 0 |
| ESP11 | 38,116 | 38,116 | 38,116 | 38,116 | 38,116 |
| ESP13 | 38,259 | 38,259 | 38,259 | 38,259 | 38,259 |
| ESP14 | 33,500 | 33,500 | 33,500 | 33,500 | 33,500 |
| ESP16 | 33,475 | 33,475 | 33,475 | 33,475 | 33,475 |
| ESP17 | 38,285 | 38,285 | 38,285 | 38,285 | 38,285 |
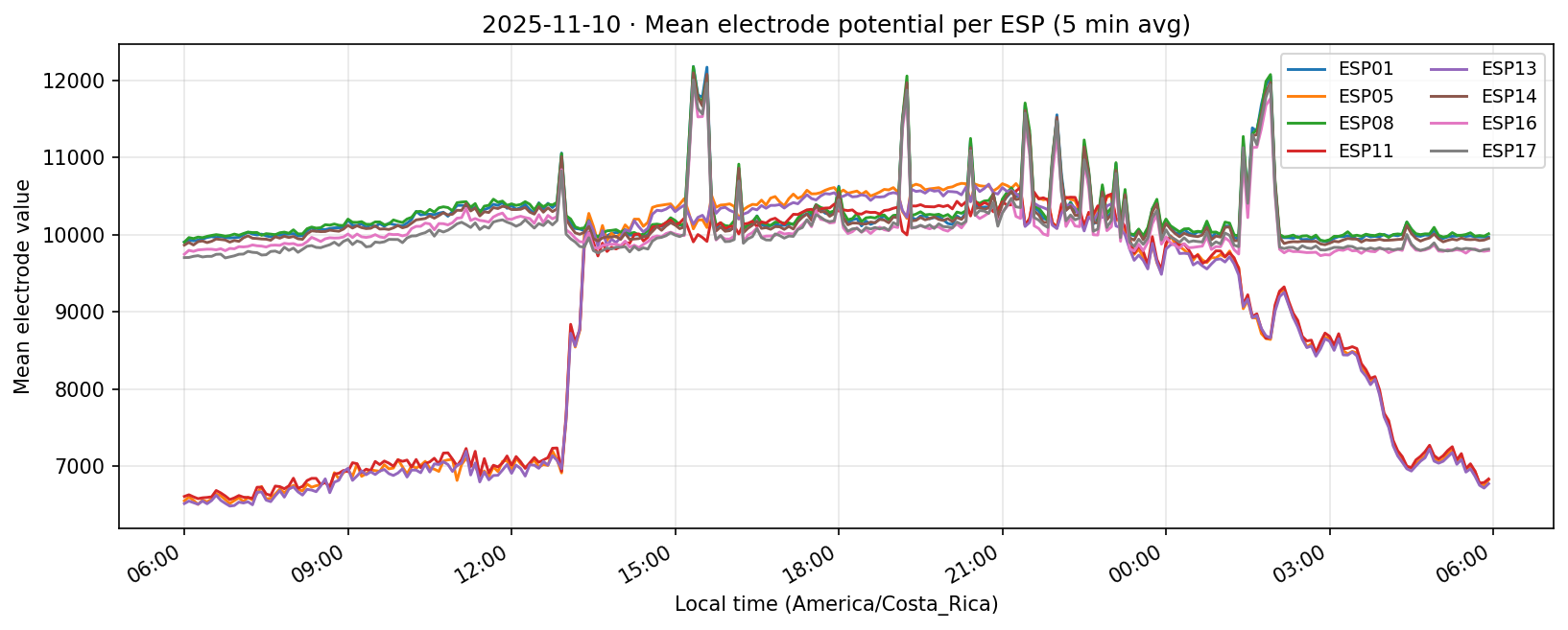
11 Nov 2025
| ESP | Total samples | a0 | a1 | a2 | a3 |
|---|---|---|---|---|---|
| ESP01 | 38,156 | 38,156 | 38,156 | 38,156 | 38,156 |
| ESP04 | 0 | 0 | 0 | 0 | 0 |
| ESP05 | 38,113 | 38,113 | 38,113 | 38,113 | 38,113 |
| ESP08 | 33,343 | 33,343 | 33,343 | 33,343 | 33,343 |
| ESP09 | 0 | 0 | 0 | 0 | 0 |
| ESP11 | 37,545 | 37,545 | 37,545 | 37,545 | 37,545 |
| ESP13 | 38,150 | 38,150 | 38,150 | 38,150 | 38,150 |
| ESP14 | 33,483 | 33,483 | 33,483 | 33,483 | 33,483 |
| ESP16 | 33,418 | 33,418 | 33,418 | 33,418 | 33,418 |
| ESP17 | 38,113 | 38,113 | 38,113 | 38,113 | 38,113 |
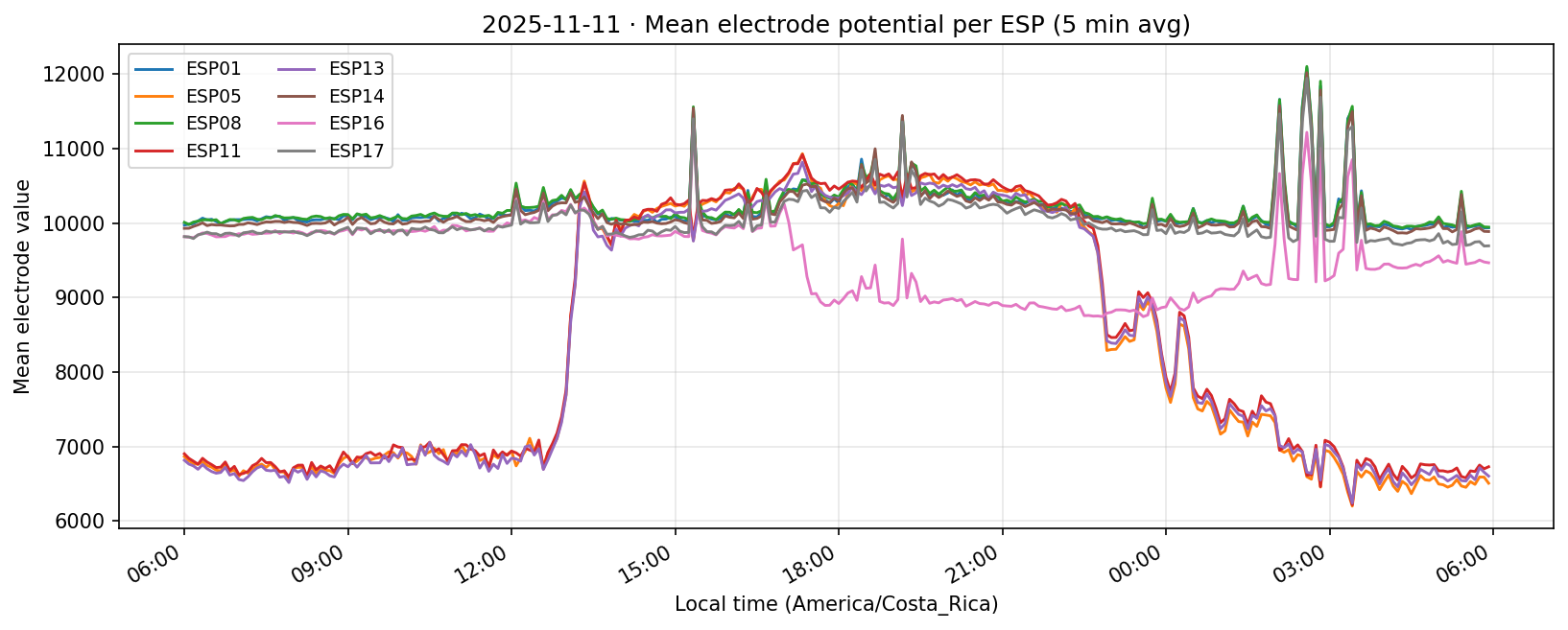
12 Nov 2025 (partial day)
| ESP | Total samples | a0 | a1 | a2 | a3 |
|---|---|---|---|---|---|
| ESP01 | 14,079 | 14,079 | 14,079 | 14,079 | 14,079 |
| ESP04 | 0 | 0 | 0 | 0 | 0 |
| ESP05 | 13,998 | 13,998 | 13,998 | 13,998 | 13,998 |
| ESP08 | 12,319 | 12,319 | 12,319 | 12,319 | 12,319 |
| ESP09 | 0 | 0 | 0 | 0 | 0 |
| ESP11 | 13,587 | 13,587 | 13,587 | 13,587 | 13,587 |
| ESP13 | 14,068 | 14,068 | 14,068 | 14,068 | 14,068 |
| ESP14 | 12,297 | 12,297 | 12,297 | 12,297 | 12,297 |
| ESP16 | 12,310 | 12,310 | 12,310 | 12,310 | 12,310 |
| ESP17 | 14,088 | 14,088 | 14,088 | 14,088 | 14,088 |
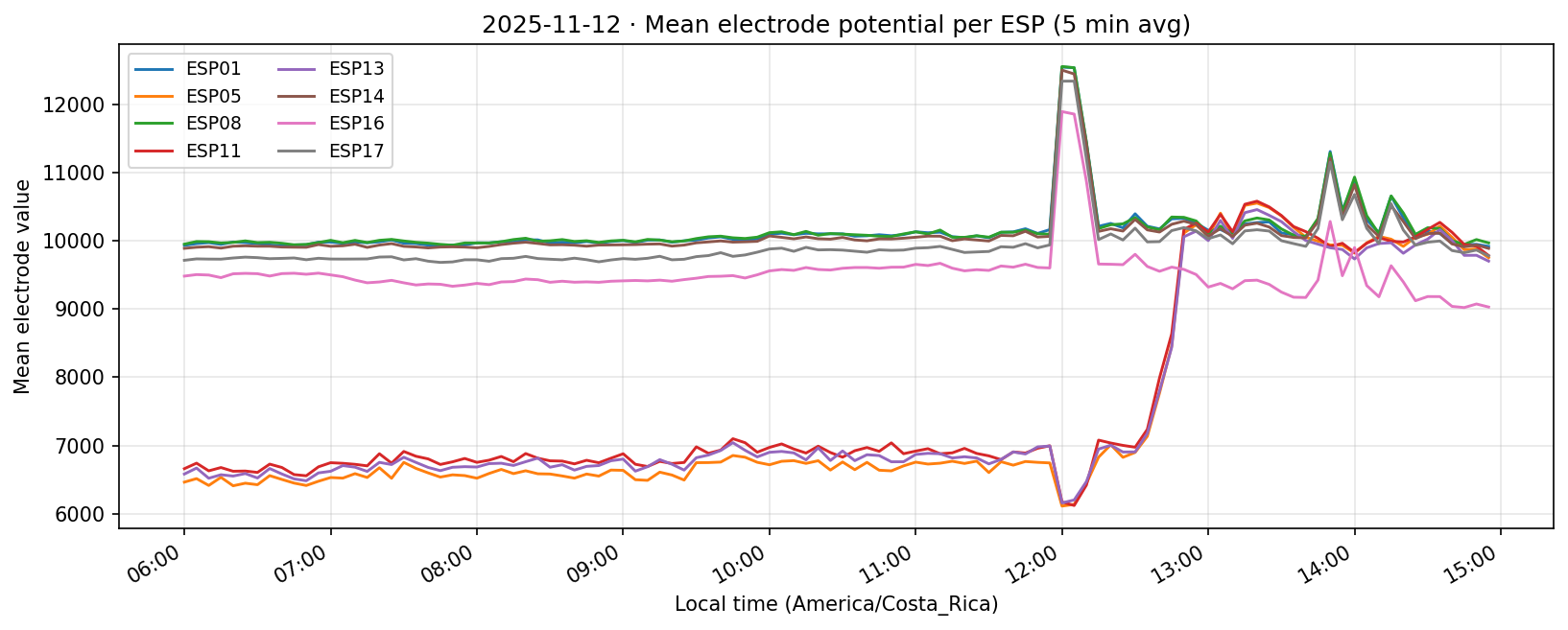
Daily counts reflect non-null readings per channel after converting to local time. Zero values indicate the device did not report for that channel on the corresponding day.
Discussion & Conclusions
What we learned. With every healthy ESP streaming cleanly through 11 Nov (18–41 k samples per channel per day), the diurnal rhythm is unmistakable: a night-time lull, a steady climb, a shallow midday dip, and an evening ramp. Just like in Experiment 001, the hourly reducer cleanly separates the devices into two clusters—Group 2 (esp01/08/14/16/17) surges ~500 counts above its dawn lows toward 19:00–20:00, while Group 1 (esp05/11/13) fires earlier, peaking near 11:00 before settling into an overnight rest. ESP16 decisively follows the Group 2 cadence now that ESP04 is offline.
Gaps we uncovered. ESPs 04 and 09 produced no data during this week, and the first/last days are partial captures (start-up/shutdown). Documenting those limits here keeps the aggregate plots honest and explains why later experiments ignore 04/09.
Where we go from here. With a complete per-ESP/per-day view established, the next step is to see what those ESP averages are actually made of. In Experiment 001B we “double click” and plot every electrode (a0–a3) separately, so we can check how sparse or uneven the contributors are before moving on to the hyphal connectivity study.
Reproducibility & Data Accessibility
- Raw CSVs:
analysis/fungal_state_mapping/outputs/experiment_001a/CSV/espXX_experiment_001a.csv. - Extraction script:
analysis/fungal_state_mapping/scripts/extract_experiment_001a_range.py. - Daily coverage summary:
analysis/fungal_state_mapping/scripts/summarize_daily_channel_counts_001a.py→analysis/fungal_state_mapping/outputs/experiment_001a/CSV/daily_channel_counts.csv. - HTML formatter:
analysis/fungal_state_mapping/scripts/print_counts_001a.py. - Per-day coverage assets:
analysis/fungal_state_mapping/scripts/generate_daily_coverage_assets.pywritesexperiments/experiment-001a/day_coverage_sections.htmlandexperiments/experiment-001a/assets/day_plots/. - Plot regeneration:
analysis/fungal_state_mapping/scripts/generate_experiment_001a_plots.pyproduces the hourly cycle and power figures above (saline assets reused from Experiment 001; wiring diagram stored atexperiments/experiment-001a/assets/wiring_patter_visulaiztion_001a.png).
References
EarthTalk Lab internal documentation on electrode calibration and saline baselines (2025).