Abstract
Motivation. We suspected the fungus cycles through electrical ΓÇ£statesΓÇ¥ (resting, feeding, growing, etc.) across the day, and that stimuli might trigger different responses depending on the state it is already in. Before naming or modelling those states, we needed to prove they exist and to see how stable the baseline is.
Approach. Using the 24ΓÇô30 Oct 2025 baseline, we averaged raw electrode readings (ESPs 04, 05, 08, 11, 13, 14, 17) into 10-minute blocks, rolled them into hourly means, and clustered the hourly curves. We then applied the exact same reducer to saline-control runs (3ΓÇô9 Oct) to check for hardware artefacts, and finally broke the fungus run back into individual days to see which hours truly contributed to the ΓÇ£average.ΓÇ¥
Results. The fungus traces exhibit a consistent dayΓÇônight rhythm and two spatially coherent clusters. That split raises the possibility that the single physical block may actually host two semi-independent mycelial ΓÇ£organismsΓÇ¥ (or at least two regions connected by too few hyphae to share state quickly). The saline control stayed largely flat, aside from a noon dip on the hottest days that we attribute to bath heating/evaporation rather than instrumentation drift. Daily coverage revealed that only 23ΓÇô25 Oct contained full data, so later hours in the aggregate trace were based on sparse or zero samples.
Conclusions & next steps. The baseline does contain repeatable states, but we need denser data (Experiment 001A) and a temperature-controlled saline repeat before we can label those states or tie stimuli to them. This report documents the processing pipeline, sanity checks, gaps discovered, and the follow-up experiments required.
Introduction
Understanding how fungal electrical signalling varies through circadian cycles is critical for correlating physiological states (“sleeping,” “feeding,” “growing,” etc.) with environmental drivers. We hypothesize that responses are state-dependent—stimulus → state → outcome—so the first question was simply: do repeatable electrical states exist over the day?
Previous EarthTalk experiments highlighted response peaks during stimulus protocols; here we characterize the inherent baseline dynamics. We focus on ESPs encircling a single substrate block, capturing absolute electrode potentials (a0ΓÇôa3) referenced to a shared ground.
To keep the narrative linear, this page follows the same order we worked through in the lab: first the hourly aggregation (Results) to look for states, then a saline sanity check (Saline control) to confirm the rhythm is biological, and finally a per-day coverage audit (Data coverage) that revealed the gaps which triggered Experiment 001A.
Materials & Methods
Data Acquisition
Raw voltage readings were logged every Γëê2 seconds by each electrode channel (a0ΓÇôa3) for ESPs 04, 05, 08, 11, 13, 14, and 17. (ESP07 had already been decommissioned before this baseline run, which is why it does not appear in the figures.) Data were exported to CSV from the InfluxDB bucket blocks_raw using the helper script push_experiment_to_notion.py for esp04 and pre-existing exports for all other ESPs.
Signal Processing
- Ten-minute block means. For each ESP, samples were partitioned into contiguous 10-minute windows. Within each window, we averaged all readings per electrode, then averaged across a0ΓÇôa3, producing one ΓÇ£mean block potentialΓÇ¥.
- Hourly aggregation. Ten-minute values whose start time fell within the same clock hour were averaged across the six-day run, yielding a multi-day hourly mean per ESP.
- State mapping (esp04). Using
analyze_fungal_states.py, we computed biological σ by subtracting saline noise in quadrature, assigned state bins (S0–S4), and produced fixed/rolling baseline visualizations with spike smoothing <3 samples.
- Take every 10-minute chunk inside an hour (00:00–00:10, 00:10–00:20, …) and average all four electrodes together for that chunk.
- Average those six chunk values to get a single number for that hour on that day.
- Repeat for each day, then average the per-day hour values to obtain the final point we plot for that hour.
- Do the same for every hour of the day so the curves show the true diurnal shape.
Software
Processing relied on Python 3.10+, Pandas, and Matplotlib. Scripts central to this study:
compute_hourly_cycle_multi_from_csv.pyΓÇö hourly aggregation & plotting from CSV exports.compute_means_10min.pyΓÇö 10-minute mean tables, min/max windows, and bin counts.analyze_fungal_states.pyΓÇö state classification for targeted windows.
Results
Global Overview

Cluster Analysis



Spatial Context

Emerging hypothesis. The two hourly clusters show up on opposite sides of the block and barely influence one another. That raised the possibility that weΓÇÖre effectively monitoring two semi-independent mycelial organisms sharing the same substrate. To test that, we plan a follow-up where we inoculate a fresh block very lightly, record its state map while the hyphae are sparse, then keep logging for several weeks as the hyphae knit the regions together. If the hourly states gradually synchronize as connectivity increases, weΓÇÖll have evidence that a minimum hyphal ΓÇ£bandwidthΓÇ¥ is required for full state transfer.
Saline Control (Hardware Baseline)
To ensure the observed diurnal structure is biological rather than an instrumentation artifact, we applied the identical aggregation pipeline to saline calibration runs collected 3ΓÇô9 October 2025. The aggregate control curve shows a modest noon dip only on the final three days (7ΓÇô9 Oct) when the uncovered bath warmed and partly evaporated; earlier days stay flat within Γëê50 counts. We therefore treat the trough as temperature-driven drift, not a fundamental property of the hardware. Raw saline CSVs live under analysis/local_workspace/data_for_professor_Adamatzky/espXX_saline_experiment_20251003_to_20251009.csv, and the derived hourly tables/plots are in analysis/local_workspace/fungal_state_mapping_outputs/saline/.




Data Coverage
Every timestamp was converted to Costa Rica local time (UTC−6) and grouped by day, yielding per-ESP sample counts and a representative trace for each date. This makes it clear which days actually contribute to the hourly averages.
Quick read. Only 23ΓÇô25 Oct retained dense coverage across all channels; afterwards the electrodes streamed reference channels only, so the daily plots flatten and the counts drop to zero, signalling the effective end of the baseline capture. That realization directly motivated Experiment 001A, which repeats the workflow on a cleaner slice (06ΓÇô12 Nov) with all ESPs online.
23 Oct 2025
| ESP | Total samples | a0 | a1 | a2 | a3 |
|---|---|---|---|---|---|
| ESP04 | 8,536 | 8,536 | 8,536 | 8,536 | 8,536 |
| ESP05 | 9,726 | 9,726 | 9,726 | 9,726 | 9,726 |
| ESP08 | 8,570 | 8,570 | 8,570 | 8,570 | 8,570 |
| ESP11 | 9,718 | 9,718 | 9,718 | 9,718 | 9,718 |
| ESP13 | 9,735 | 9,735 | 9,735 | 9,735 | 9,735 |
| ESP14 | 8,605 | 8,605 | 8,605 | 8,605 | 8,605 |
| ESP17 | 9,706 | 9,706 | 9,706 | 9,706 | 9,706 |
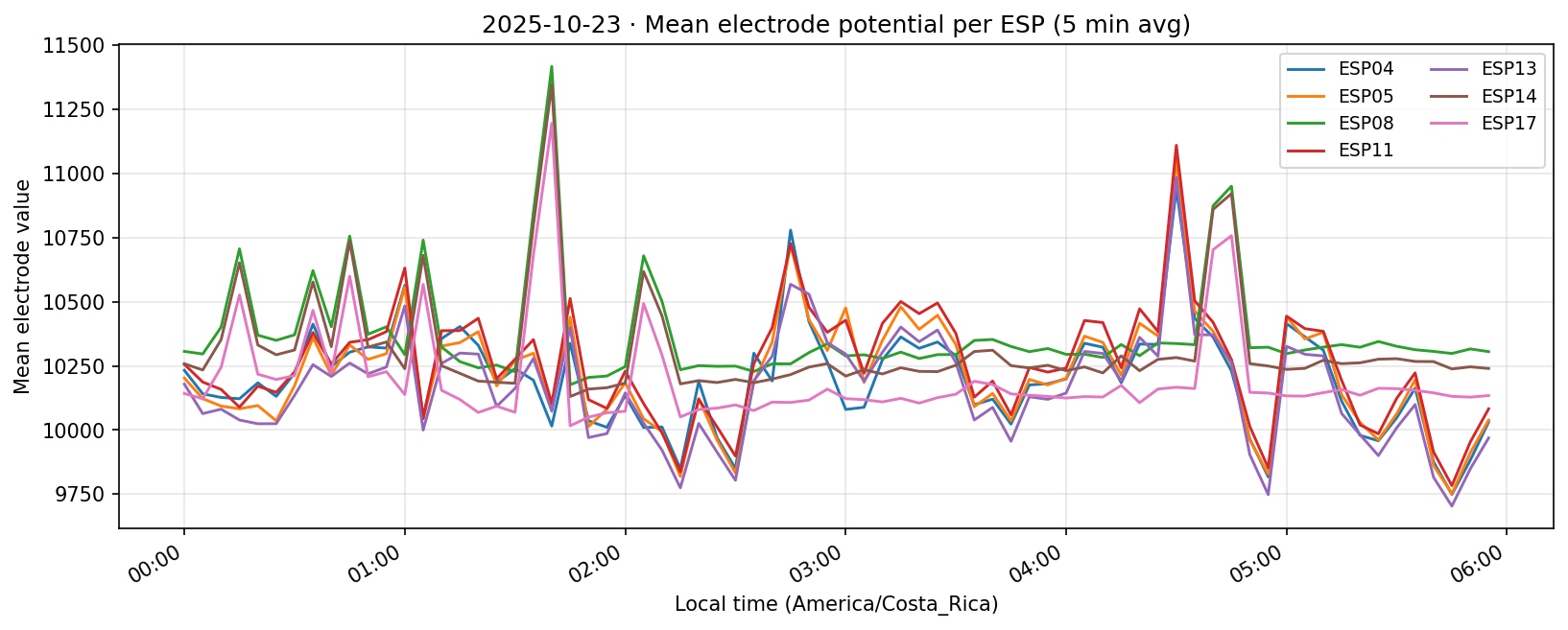
24 Oct 2025
| ESP | Total samples | a0 | a1 | a2 | a3 |
|---|---|---|---|---|---|
| ESP04 | 33,162 | 33,162 | 33,162 | 33,162 | 33,162 |
| ESP05 | 37,051 | 37,051 | 37,051 | 37,051 | 37,051 |
| ESP08 | 33,196 | 33,196 | 33,196 | 33,196 | 33,196 |
| ESP11 | 37,709 | 37,709 | 37,709 | 37,709 | 37,709 |
| ESP13 | 37,681 | 37,681 | 37,681 | 37,681 | 37,681 |
| ESP14 | 33,316 | 33,316 | 33,316 | 33,316 | 33,316 |
| ESP17 | 37,668 | 37,668 | 37,668 | 37,668 | 37,668 |
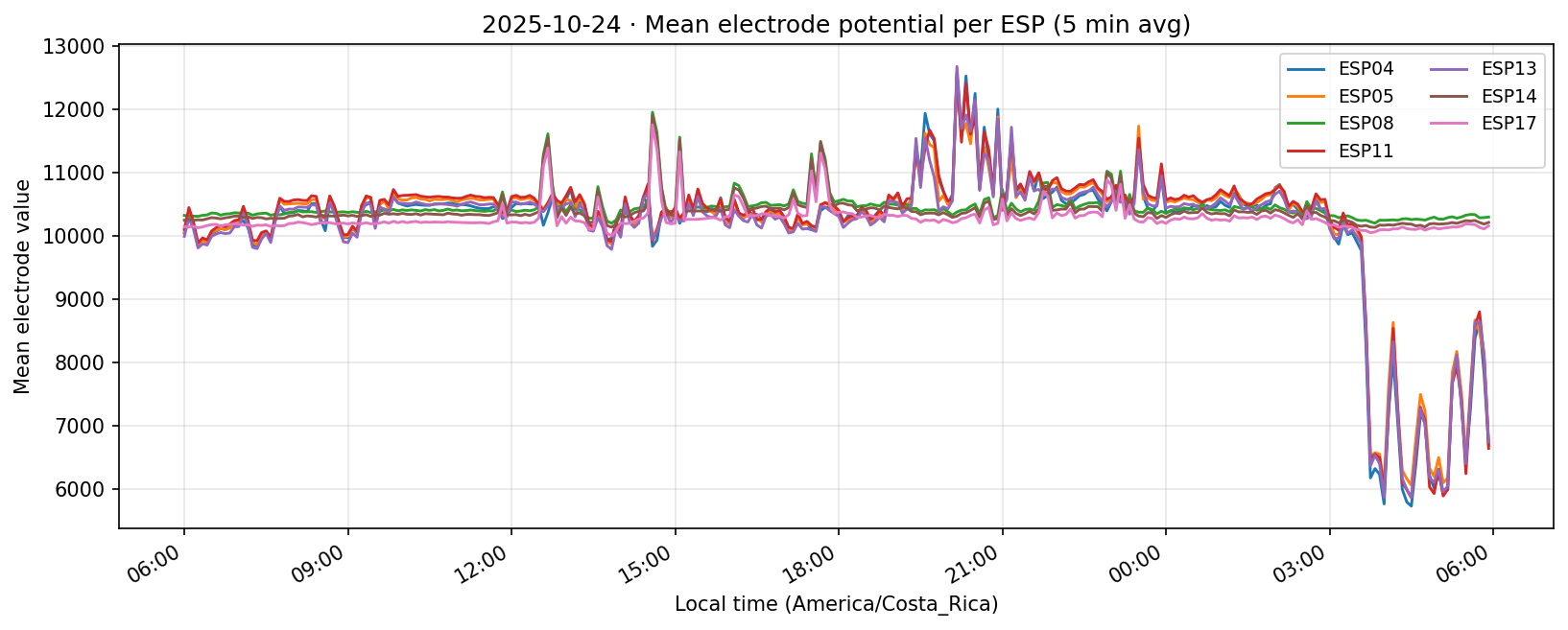
25 Oct 2025
| ESP | Total samples | a0 | a1 | a2 | a3 |
|---|---|---|---|---|---|
| ESP04 | 21,938 | 15,431 | 15,431 | 15,431 | 15,431 |
| ESP05 | 24,890 | 17,487 | 17,487 | 17,487 | 17,487 |
| ESP08 | 21,964 | 15,446 | 15,446 | 15,446 | 15,446 |
| ESP11 | 24,936 | 17,533 | 17,533 | 17,533 | 17,533 |
| ESP13 | 24,988 | 17,575 | 17,575 | 17,575 | 17,575 |
| ESP14 | 22,075 | 15,539 | 15,539 | 15,539 | 15,539 |
| ESP17 | 24,944 | 17,556 | 17,556 | 17,556 | 17,556 |
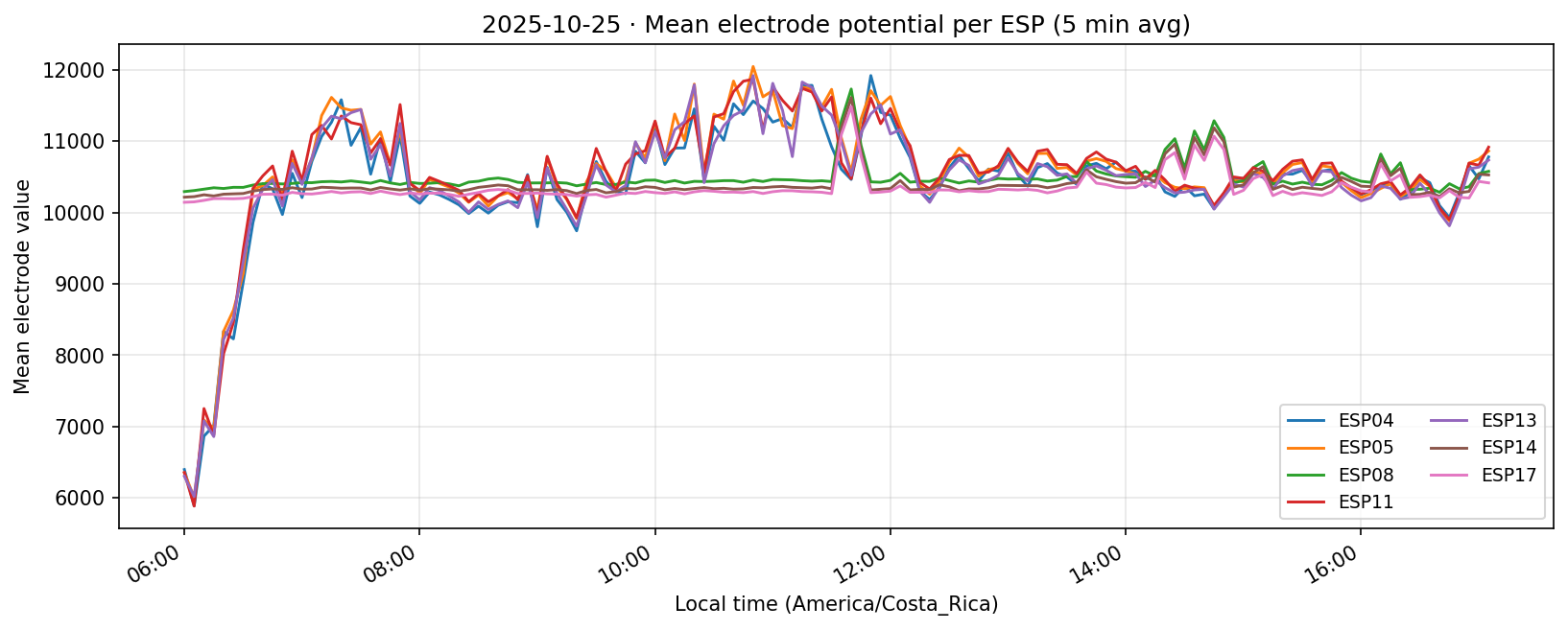
26 Oct 2025
| ESP | Total samples | a0 | a1 | a2 | a3 |
|---|---|---|---|---|---|
| ESP04 | 26,886 | 0 | 0 | 0 | 0 |
| ESP05 | 30,620 | 0 | 0 | 0 | 0 |
| ESP08 | 26,997 | 0 | 0 | 0 | 0 |
| ESP11 | 30,579 | 0 | 0 | 0 | 0 |
| ESP13 | 30,662 | 0 | 0 | 0 | 0 |
| ESP14 | 27,001 | 0 | 0 | 0 | 0 |
| ESP17 | 30,640 | 0 | 0 | 0 | 0 |
27 Oct 2025
| ESP | Total samples | a0 | a1 | a2 | a3 |
|---|---|---|---|---|---|
| ESP04 | 0 | 0 | 0 | 0 | 0 |
| ESP05 | 37,179 | 0 | 0 | 0 | 0 |
| ESP08 | 32,646 | 0 | 0 | 0 | 0 |
| ESP11 | 37,007 | 0 | 0 | 0 | 0 |
| ESP13 | 37,292 | 0 | 0 | 0 | 0 |
| ESP14 | 32,695 | 0 | 0 | 0 | 0 |
| ESP17 | 37,255 | 0 | 0 | 0 | 0 |
28 Oct 2025
| ESP | Total samples | a0 | a1 | a2 | a3 |
|---|---|---|---|---|---|
| ESP04 | 0 | 0 | 0 | 0 | 0 |
| ESP05 | 37,641 | 0 | 0 | 0 | 0 |
| ESP08 | 32,900 | 0 | 0 | 0 | 0 |
| ESP11 | 37,580 | 0 | 0 | 0 | 0 |
| ESP13 | 37,595 | 0 | 0 | 0 | 0 |
| ESP14 | 32,966 | 0 | 0 | 0 | 0 |
| ESP17 | 37,578 | 0 | 0 | 0 | 0 |
29 Oct 2025
| ESP | Total samples | a0 | a1 | a2 | a3 |
|---|---|---|---|---|---|
| ESP04 | 0 | 0 | 0 | 0 | 0 |
| ESP05 | 37,883 | 0 | 0 | 0 | 0 |
| ESP08 | 33,197 | 0 | 0 | 0 | 0 |
| ESP11 | 37,717 | 0 | 0 | 0 | 0 |
| ESP13 | 37,946 | 0 | 0 | 0 | 0 |
| ESP14 | 33,207 | 0 | 0 | 0 | 0 |
| ESP17 | 37,918 | 0 | 0 | 0 | 0 |
30 Oct 2025
| ESP | Total samples | a0 | a1 | a2 | a3 |
|---|---|---|---|---|---|
| ESP04 | 0 | 0 | 0 | 0 | 0 |
| ESP05 | 28,115 | 0 | 0 | 0 | 0 |
| ESP08 | 24,542 | 0 | 0 | 0 | 0 |
| ESP11 | 28,174 | 0 | 0 | 0 | 0 |
| ESP13 | 28,084 | 0 | 0 | 0 | 0 |
| ESP14 | 24,601 | 0 | 0 | 0 | 0 |
| ESP17 | 28,098 | 0 | 0 | 0 | 0 |
Daily counts reflect non-null readings per channel after converting to Costa Rica local time. Zero values indicate the channel did not report that day.
Discussion & Conclusions
What we learned. The hourly reducer shows that the fungus naturally settles into a night-time lull, climbs through the morning, dips slightly around noon, then ramps again toward evening. Two spatial clusters behave almost like twins, which hints that location (moisture, shade, airflow) influences the baseline voltage.
Why the saline check matters. Running the same math on the saline bath confirmed that the instrumentation itself stays essentially flat; the only wiggle is the noon dip on 7ΓÇô9 Oct when the bath warmed and evaporated. That gives us confidence that the bigger swings in the fungus traces are biological and not an ADC quirk.
Gaps we uncovered. Once we inspected the daily counts we saw that most channels went silent after 25 Oct. Instead of quietly averaging in hours with no true data, we documented the gap here and spun up Experiment 001A to redo the baseline with solid coverage.
Where we go from here. The concrete follow-ups (canonical baseline, saline rerun, hyphal-connectivity test) are listed in the Future Experiments section so they stay visible as active work items.
Future Experiments
- Baseline reboot (Experiment 001A). Use the 06ΓÇô12 Nov capture (all channels healthy) as the canonical baseline going forward, and rerun the hourly clustering on that dataset to confirm the spatial groupings hold when coverage is complete.
- Temperature-controlled saline repeat. Re-run the saline baseline with the bath in a thermostated enclosure (or at least covered and continuously topped off) to confirm that the noon dip truly tracks ambient heating/evaporation. Capturing synchronized ambient temperature alongside the control will let us regress out residual drift and tighten future normalization steps.
- Hyphal connectivity / state-transfer study. Start with a lightly inoculated block, log per-hour states while regions are poorly connected, then let the hyphae knit together and monitor whether the two electrical clusters synchronize. This will tell us whether a minimum hyphal ΓÇ£bandwidthΓÇ¥ is required for full state sharing.
Reproducibility & Data Accessibility
- Raw CSVs:
analysis/fungal_state_mapping/outputs/experiment_001/CSV/espXX_experiment_001.csv. - Extraction script:
analysis/fungal_state_mapping/scripts/extract_experiment_001_range.py. - Hourly aggregation:
compute_hourly_cycle_multi_from_csv.py,compute_means_10min.py,analyze_fungal_states.py,query_influx_mean_std.py. - Daily coverage assets:
analysis/fungal_state_mapping/scripts/generate_daily_coverage_assets.pywritesexperiments/experiment-001/day_coverage_sections.htmlandexperiments/experiment-001/assets/day_plots/. - Legacy state plots:
fixed_baseline_states.png,rolling_baseline_states.png(ESP04 focus).
References
EarthTalk Lab internal documentation on electrode calibration and saline baselines (2025).